是否有人对如何将 Sparklyr 的 ml_decision_tree_classifier、ml_gbt_classifier 或 ml_random_forest_classifier 模型中的树信息转换为 a.) 其他 R 树相关库可以理解的格式以及(最终)b.) 可视化有任何建议树木的非技术消耗?这包括从矢量汇编器期间生成的替换字符串索引值转换回实际特征名称的能力。
以下代码是从 a sparklyr blog post 随意复制的为了提供示例:
library(sparklyr)
library(dplyr)
# If needed, install Spark locally via `spark_install()`
sc <- spark_connect(master = "local")
iris_tbl <- copy_to(sc, iris)
# split the data into train and validation sets
iris_data <- iris_tbl %>%
sdf_partition(train = 2/3, validation = 1/3, seed = 123)
iris_pipeline <- ml_pipeline(sc) %>%
ft_dplyr_transformer(
iris_data$train %>%
mutate(Sepal_Length = log(Sepal_Length),
Sepal_Width = Sepal_Width ^ 2)
) %>%
ft_string_indexer("Species", "label")
iris_pipeline_model <- iris_pipeline %>%
ml_fit(iris_data$train)
iris_vector_assembler <- ft_vector_assembler(
sc,
input_cols = setdiff(colnames(iris_data$train), "Species"),
output_col = "features"
)
random_forest <- ml_random_forest_classifier(sc,features_col = "features")
# obtain the labels from the fitted StringIndexerModel
iris_labels <- iris_pipeline_model %>%
ml_stage("string_indexer") %>%
ml_labels()
# IndexToString will convert the predicted numeric values back to class labels
iris_index_to_string <- ft_index_to_string(sc, "prediction", "predicted_label",
labels = iris_labels)
# construct a pipeline with these stages
iris_prediction_pipeline <- ml_pipeline(
iris_pipeline, # pipeline from previous section
iris_vector_assembler,
random_forest,
iris_index_to_string
)
# fit to data and make some predictions
iris_prediction_model <- iris_prediction_pipeline %>%
ml_fit(iris_data$train)
iris_predictions <- iris_prediction_model %>%
ml_transform(iris_data$validation)
iris_predictions %>%
select(Species, label:predicted_label) %>%
glimpse()
根据 here 的建议进行反复试验我能够以“if/else”格式打印出底层决策树的公式,并将其转换为字符串:
model_stage <- iris_prediction_model$stages[[3]]
spark_jobj(model_stage) %>% invoke(., "toDebugString") %>% cat()
##print out below##
RandomForestClassificationModel (uid=random_forest_classifier_5c6a1934c8e) with 20 trees
Tree 0 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 4.95)
If (feature 3 <= 1.65)
Predict: 0.0
Else (feature 3 > 1.65)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Else (feature 2 > 4.95)
If (feature 2 <= 5.05)
If (feature 1 <= 6.505000000000001)
Predict: 2.0
Else (feature 1 > 6.505000000000001)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 1 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.75)
If (feature 1 <= 5.0649999999999995)
If (feature 3 <= 1.05)
Predict: 0.0
Else (feature 3 > 1.05)
If (feature 0 <= 1.8000241202036602)
Predict: 2.0
Else (feature 0 > 1.8000241202036602)
Predict: 0.0
Else (feature 1 > 5.0649999999999995)
If (feature 0 <= 1.8000241202036602)
Predict: 0.0
Else (feature 0 > 1.8000241202036602)
If (feature 2 <= 5.05)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 2 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 0 <= 1.7664051342320237)
Predict: 0.0
Else (feature 0 > 1.7664051342320237)
If (feature 3 <= 1.45)
If (feature 2 <= 4.85)
Predict: 0.0
Else (feature 2 > 4.85)
Predict: 2.0
Else (feature 3 > 1.45)
If (feature 3 <= 1.65)
If (feature 1 <= 8.125)
Predict: 2.0
Else (feature 1 > 8.125)
Predict: 0.0
Else (feature 3 > 1.65)
Predict: 2.0
Tree 3 (weight 1.0):
If (feature 0 <= 1.6675287895788053)
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
Predict: 0.0
Else (feature 0 > 1.6675287895788053)
If (feature 3 <= 1.75)
If (feature 3 <= 1.55)
If (feature 1 <= 7.025)
If (feature 2 <= 4.55)
Predict: 0.0
Else (feature 2 > 4.55)
Predict: 2.0
Else (feature 1 > 7.025)
Predict: 0.0
Else (feature 3 > 1.55)
If (feature 2 <= 5.05)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 4 (weight 1.0):
If (feature 2 <= 4.85)
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
Predict: 0.0
Else (feature 2 > 4.85)
If (feature 2 <= 5.05)
If (feature 0 <= 1.8484238118815566)
Predict: 2.0
Else (feature 0 > 1.8484238118815566)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 5 (weight 1.0):
If (feature 2 <= 1.65)
Predict: 1.0
Else (feature 2 > 1.65)
If (feature 3 <= 1.65)
If (feature 0 <= 1.8325494627242664)
Predict: 0.0
Else (feature 0 > 1.8325494627242664)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Else (feature 3 > 1.65)
Predict: 2.0
Tree 6 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 5.05)
If (feature 3 <= 1.75)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 7 (weight 1.0):
If (feature 3 <= 0.55)
Predict: 1.0
Else (feature 3 > 0.55)
If (feature 3 <= 1.65)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
Predict: 2.0
Else (feature 3 > 1.65)
If (feature 2 <= 4.85)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Else (feature 2 > 4.85)
Predict: 2.0
Tree 8 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.85)
If (feature 2 <= 4.85)
Predict: 0.0
Else (feature 2 > 4.85)
If (feature 0 <= 1.8794359129669855)
Predict: 2.0
Else (feature 0 > 1.8794359129669855)
If (feature 3 <= 1.55)
Predict: 0.0
Else (feature 3 > 1.55)
Predict: 0.0
Else (feature 3 > 1.85)
Predict: 2.0
Tree 9 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Tree 10 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
If (feature 2 <= 5.05)
If (feature 3 <= 1.55)
Predict: 2.0
Else (feature 3 > 1.55)
If (feature 3 <= 1.75)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 11 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 2 <= 5.05)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 3 <= 1.75)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 12 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.75)
If (feature 3 <= 1.35)
Predict: 0.0
Else (feature 3 > 1.35)
If (feature 0 <= 1.695573522904327)
Predict: 0.0
Else (feature 0 > 1.695573522904327)
If (feature 1 <= 8.125)
Predict: 2.0
Else (feature 1 > 8.125)
Predict: 0.0
Else (feature 3 > 1.75)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Tree 13 (weight 1.0):
If (feature 3 <= 0.55)
Predict: 1.0
Else (feature 3 > 0.55)
If (feature 2 <= 4.95)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 0 <= 1.8000241202036602)
If (feature 1 <= 9.305)
Predict: 2.0
Else (feature 1 > 9.305)
Predict: 0.0
Else (feature 0 > 1.8000241202036602)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Tree 14 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 3 <= 1.65)
If (feature 3 <= 1.45)
Predict: 0.0
Else (feature 3 > 1.45)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Else (feature 3 > 1.65)
If (feature 0 <= 1.7833559100698644)
If (feature 0 <= 1.7664051342320237)
Predict: 2.0
Else (feature 0 > 1.7664051342320237)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Tree 15 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 3 <= 1.75)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
If (feature 1 <= 8.125)
Predict: 2.0
Else (feature 1 > 8.125)
If (feature 0 <= 1.9095150692894909)
Predict: 0.0
Else (feature 0 > 1.9095150692894909)
Predict: 2.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 16 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 0 <= 1.7491620461964392)
Predict: 0.0
Else (feature 0 > 1.7491620461964392)
If (feature 3 <= 1.75)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 0 <= 1.8164190316151556)
Predict: 2.0
Else (feature 0 > 1.8164190316151556)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 17 (weight 1.0):
If (feature 0 <= 1.695573522904327)
If (feature 2 <= 1.65)
Predict: 1.0
Else (feature 2 > 1.65)
Predict: 0.0
Else (feature 0 > 1.695573522904327)
If (feature 2 <= 4.75)
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 3 <= 1.75)
If (feature 1 <= 5.0649999999999995)
Predict: 2.0
Else (feature 1 > 5.0649999999999995)
If (feature 3 <= 1.65)
Predict: 0.0
Else (feature 3 > 1.65)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 18 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.65)
Predict: 0.0
Else (feature 3 > 1.65)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Tree 19 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 4.95)
If (feature 1 <= 8.705)
Predict: 0.0
Else (feature 1 > 8.705)
If (feature 2 <= 4.85)
Predict: 0.0
Else (feature 2 > 4.85)
If (feature 0 <= 1.8164190316151556)
Predict: 2.0
Else (feature 0 > 1.8164190316151556)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
正如您所看到的,这种格式对于传递到我见过的可视化决策 TreeMap 形的众多漂亮方法之一来说并不是最佳选择(例如 revolution analytics 或 statmethods )
最佳答案
从今天开始(Spark 2.4.0 版本已经批准并等待官方公告),您最好的选择*,在不涉及复杂的第 3 方工具(例如您可以看看 MLeap)的情况下,可能是 save the model并回读 the specification :
ml_stage(iris_prediction_model, "random_forest") %>%
ml_save("/tmp/model")
rf_spec <- spark_read_parquet(sc, "rf", "/tmp/model/data/")
结果将是具有以下架构的 Spark DataFrame:
rf_spec %>%
spark_dataframe() %>%
invoke("schema") %>% invoke("treeString") %>%
cat(sep = "\n")
root
|-- treeID: integer (nullable = true)
|-- nodeData: struct (nullable = true)
| |-- id: integer (nullable = true)
| |-- prediction: double (nullable = true)
| |-- impurity: double (nullable = true)
| |-- impurityStats: array (nullable = true)
| | |-- element: double (containsNull = true)
| |-- gain: double (nullable = true)
| |-- leftChild: integer (nullable = true)
| |-- rightChild: integer (nullable = true)
| |-- split: struct (nullable = true)
| | |-- featureIndex: integer (nullable = true)
| | |-- leftCategoriesOrThreshold: array (nullable = true)
| | | |-- element: double (containsNull = true)
| | |-- numCategories: integer (nullable = true)
提供有关所有节点和拆分的信息。
可以使用列元数据检索特征映射:
meta <- iris_predictions %>%
select(features) %>%
spark_dataframe() %>%
invoke("schema") %>% invoke("apply", 0L) %>%
invoke("metadata") %>%
invoke("getMetadata", "ml_attr") %>%
invoke("getMetadata", "attrs") %>%
invoke("json") %>%
jsonlite::fromJSON() %>%
dplyr::bind_rows() %>%
copy_to(sc, .) %>%
rename(featureIndex = idx)
meta
# Source: spark<?> [?? x 2]
featureIndex name
* <int> <chr>
1 0 Sepal_Length
2 1 Sepal_Width
3 2 Petal_Length
4 3 Petal_Width
以及您已检索到的标签映射:
labels <- tibble(prediction = seq_along(iris_labels) - 1, label = iris_labels) %>%
copy_to(sc, .)
最后你可以将所有这些结合起来:
full_rf_spec <- rf_spec %>%
spark_dataframe() %>%
invoke("selectExpr", list("treeID", "nodeData.*", "nodeData.split.*")) %>%
sdf_register() %>%
select(-split, -impurityStats) %>%
left_join(meta, by = "featureIndex") %>%
left_join(labels, by = "prediction")
full_rf_spec
# Source: spark<?> [?? x 12]
treeID id prediction impurity gain leftChild rightChild featureIndex
* <int> <int> <dbl> <dbl> <dbl> <int> <int> <int>
1 0 0 1 0.636 0.379 1 2 2
2 0 1 1 0 -1 -1 -1 -1
3 0 2 0 0.440 0.367 3 8 2
4 0 3 0 0.0555 0.0269 4 5 3
5 0 4 0 0 -1 -1 -1 -1
6 0 5 0 0.5 0.5 6 7 0
7 0 6 0 0 -1 -1 -1 -1
8 0 7 2 0 -1 -1 -1 -1
9 0 8 2 0.111 0.0225 9 12 2
10 0 9 2 0.375 0.375 10 11 1
# ... with more rows, and 4 more variables: leftCategoriesOrThreshold <list>,
# numCategories <int>, name <chr>, label <chr>
由treeID收集和分隔,应该提供足够的信息**来模仿树状对象(您可以通过检查 rpart::rpart.object 文档和/或unclassrpart 模型。tree::tree 需要较少的工作,但其绘图实用程序远非令人印象深刻),并构建一个不错的绘图。
另一种方法是使用 Sparklyr2PMML 将数据导出到 PMML并使用这个表示。
您还可以查看How do I visualise / plot a decision tree in Apache Spark (PySpark 1.4.1)?这建议第三方Python包来解决同样的问题。
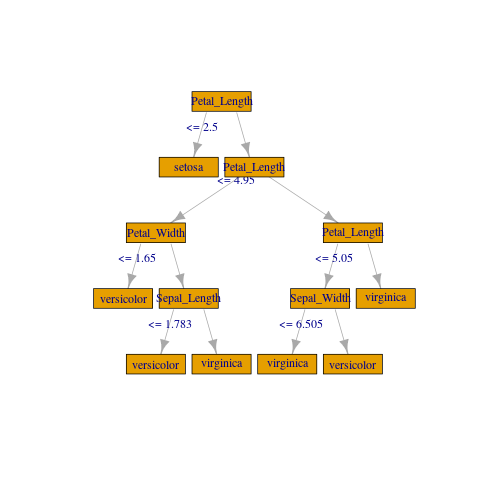
如果您不需要任何花哨的东西,您可以使用 igraph 创建一个粗略的绘图:
library(igraph)
gframe <- full_rf_spec %>%
filter(treeID == 0) %>% # Take the first tree
mutate(
leftCategoriesOrThreshold = ifelse(
size(leftCategoriesOrThreshold) == 1,
# Continuous variable case
concat("<= ", round(concat_ws("", leftCategoriesOrThreshold), 3)),
# Categorical variable case. Decoding variables might be involved
# but can be achieved if needed, using column metadata or indexer labels
concat("in {", concat_ws(",", leftCategoriesOrThreshold), "}")
),
name = coalesce(name, label)) %>%
select(
id, label, impurity, gain,
leftChild, rightChild, leftCategoriesOrThreshold, name) %>%
collect()
vertices <- gframe %>% rename(label = name, name = id)
edges <- gframe %>%
transmute(from = id, to = leftChild, label = leftCategoriesOrThreshold) %>%
union_all(gframe %>% select(from = id, to = rightChild)) %>%
filter(to != -1)
g <- igraph::graph_from_data_frame(edges, vertices = vertices)
plot(
g, layout = layout_as_tree(g, root = c(1)),
vertex.shape = "rectangle", vertex.size = 45)
* 它应该在不久的将来得到改进,通过新引入的与格式无关的 ML writer API(它已经支持选定模型的 PMML writer。希望新的模型和格式将会随之而来)。
** 如果您使用分类特征,您可能需要将 leftCategoriesOrThreshold 映射到相应的索引级别。
如果特征向量包含分类变量,则 jsonlite::fromJSON() 的输出将包含 nominal 组。例如,如果您的索引列 foo 具有三个级别,并在第一个位置组装,则它将如下所示:
$nominal
vals idx name
1 a, b, c 1 foo
其中 vals 列是可变长度向量的列表。
length(meta$nominal$vals[[1]])
[1] 3
标签对应于该结构的索引,因此在示例中:
a的标签为 0.0(不是说标签是 double float ,且从 0.0 开始编号)b的标签为 1.0
依此类推,如果您使用 leftCategoriesOrThreshold 进行分割,等于 c(0.0, 2.0),则意味着分割在标签上 {"a”,“c”}。
另请注意,如果存在分类数据,您可能必须在调用 copy_to 之前对其进行处理 - 目前看来它不支持复杂字段。
在 Spark <= 2.3 中,您将必须使用 R 代码进行映射(在本地结构上,一些 purrr 应该就可以了)。在 Spark 2.4 中(sparklyr AFAIK 尚不支持),使用 Spark 的 JSON 读取器直接读取元数据并使用其高阶函数进行映射可能会更容易。
关于r - 从 Sparklyr 中提取模型树并进行可视化,我们在Stack Overflow上找到一个类似的问题: https://stackoverflow.com/questions/53123840/