我正在使用 numpy 和 scipy 处理一些用 CCD 相机拍摄的图像。这些图像有许多具有非常大(或小)值的热(和死)像素。这些会干扰其他图像处理,因此需要将其删除。不幸的是,尽管一些像素停留在 0 或 255,并且在所有图像中始终处于相同的值,但仍有一些像素暂时停留在其他值几分钟(数据跨度很长时间)。
我想知道是否有一种方法可以识别(和删除)已经在 python 中实现的热像素。如果没有,我想知道这样做的有效方法是什么。通过将它们与相邻像素进行比较,热/坏像素相对容易识别。我可以看到编写一个循环来查看每个像素,将其值与其 8 个最近邻居的值进行比较。或者,使用某种卷积来生成更平滑的图像,然后从包含热像素的图像中减去它似乎更好,使它们更容易识别。
我已经在下面的代码中尝试了这种“模糊方法”,它工作正常,但我怀疑它是否是最快的。此外,它在图像的边缘变得困惑(可能是因为 gaussian_filter 函数正在进行卷积并且卷积在边缘附近变得奇怪)。那么,有没有更好的方法来解决这个问题?
示例代码:
import numpy as np
import matplotlib.pyplot as plt
import scipy.ndimage
plt.figure(figsize=(8,4))
ax1 = plt.subplot(121)
ax2 = plt.subplot(122)
#make a sample image
x = np.linspace(-5,5,200)
X,Y = np.meshgrid(x,x)
Z = 255*np.cos(np.sqrt(x**2 + Y**2))**2
for i in range(0,11):
#Add some hot pixels
Z[np.random.randint(low=0,high=199),np.random.randint(low=0,high=199)]= np.random.randint(low=200,high=255)
#and dead pixels
Z[np.random.randint(low=0,high=199),np.random.randint(low=0,high=199)]= np.random.randint(low=0,high=10)
#Then plot it
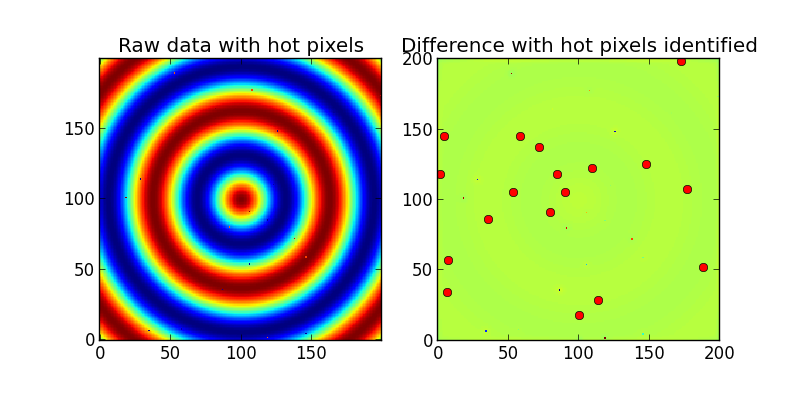
ax1.set_title('Raw data with hot pixels')
ax1.imshow(Z,interpolation='nearest',origin='lower')
#Now we try to find the hot pixels
blurred_Z = scipy.ndimage.gaussian_filter(Z, sigma=2)
difference = Z - blurred_Z
ax2.set_title('Difference with hot pixels identified')
ax2.imshow(difference,interpolation='nearest',origin='lower')
threshold = 15
hot_pixels = np.nonzero((difference>threshold) | (difference<-threshold))
#Don't include the hot pixels that we found near the edge:
count = 0
for y,x in zip(hot_pixels[0],hot_pixels[1]):
if (x != 0) and (x != 199) and (y != 0) and (y != 199):
ax2.plot(x,y,'ro')
count += 1
print 'Detected %i hot/dead pixels out of 20.'%count
ax2.set_xlim(0,200); ax2.set_ylim(0,200)
plt.show()
输出:
最佳答案
基本上,我认为处理热像素最快的方法就是使用 size=2 的中值滤波器。然后,噗,你的热像素消失了,你也消除了来自相机的各种其他高频传感器噪音。
如果您真的只想删除热像素,那么您可以像我在问题中所做的那样从原始图像中减去中值滤波器,然后仅将这些值替换为中值滤波图像中的值。这在边缘处效果不佳,因此如果您可以忽略沿边缘的像素,那么这将使事情变得容易得多。
如果要处理边缘,可以使用下面的代码。然而,它并不是最快的:
import numpy as np
import matplotlib.pyplot as plt
import scipy.ndimage
plt.figure(figsize=(10,5))
ax1 = plt.subplot(121)
ax2 = plt.subplot(122)
#make some sample data
x = np.linspace(-5,5,200)
X,Y = np.meshgrid(x,x)
Z = 100*np.cos(np.sqrt(x**2 + Y**2))**2 + 50
np.random.seed(1)
for i in range(0,11):
#Add some hot pixels
Z[np.random.randint(low=0,high=199),np.random.randint(low=0,high=199)]= np.random.randint(low=200,high=255)
#and dead pixels
Z[np.random.randint(low=0,high=199),np.random.randint(low=0,high=199)]= np.random.randint(low=0,high=10)
#And some hot pixels in the corners and edges
Z[0,0] =255
Z[-1,-1] =255
Z[-1,0] =255
Z[0,-1] =255
Z[0,100] =255
Z[-1,100]=255
Z[100,0] =255
Z[100,-1]=255
#Then plot it
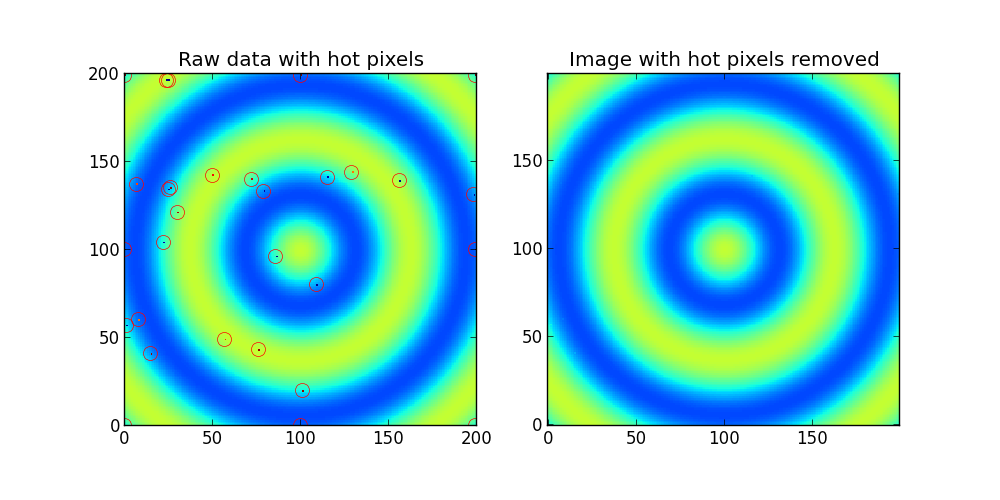
ax1.set_title('Raw data with hot pixels')
ax1.imshow(Z,interpolation='nearest',origin='lower')
def find_outlier_pixels(data,tolerance=3,worry_about_edges=True):
#This function finds the hot or dead pixels in a 2D dataset.
#tolerance is the number of standard deviations used to cutoff the hot pixels
#If you want to ignore the edges and greatly speed up the code, then set
#worry_about_edges to False.
#
#The function returns a list of hot pixels and also an image with with hot pixels removed
from scipy.ndimage import median_filter
blurred = median_filter(Z, size=2)
difference = data - blurred
threshold = 10*np.std(difference)
#find the hot pixels, but ignore the edges
hot_pixels = np.nonzero((np.abs(difference[1:-1,1:-1])>threshold) )
hot_pixels = np.array(hot_pixels) + 1 #because we ignored the first row and first column
fixed_image = np.copy(data) #This is the image with the hot pixels removed
for y,x in zip(hot_pixels[0],hot_pixels[1]):
fixed_image[y,x]=blurred[y,x]
if worry_about_edges == True:
height,width = np.shape(data)
###Now get the pixels on the edges (but not the corners)###
#left and right sides
for index in range(1,height-1):
#left side:
med = np.median(data[index-1:index+2,0:2])
diff = np.abs(data[index,0] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[index],[0]] ))
fixed_image[index,0] = med
#right side:
med = np.median(data[index-1:index+2,-2:])
diff = np.abs(data[index,-1] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[index],[width-1]] ))
fixed_image[index,-1] = med
#Then the top and bottom
for index in range(1,width-1):
#bottom:
med = np.median(data[0:2,index-1:index+2])
diff = np.abs(data[0,index] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[0],[index]] ))
fixed_image[0,index] = med
#top:
med = np.median(data[-2:,index-1:index+2])
diff = np.abs(data[-1,index] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[height-1],[index]] ))
fixed_image[-1,index] = med
###Then the corners###
#bottom left
med = np.median(data[0:2,0:2])
diff = np.abs(data[0,0] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[0],[0]] ))
fixed_image[0,0] = med
#bottom right
med = np.median(data[0:2,-2:])
diff = np.abs(data[0,-1] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[0],[width-1]] ))
fixed_image[0,-1] = med
#top left
med = np.median(data[-2:,0:2])
diff = np.abs(data[-1,0] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[height-1],[0]] ))
fixed_image[-1,0] = med
#top right
med = np.median(data[-2:,-2:])
diff = np.abs(data[-1,-1] - med)
if diff>threshold:
hot_pixels = np.hstack(( hot_pixels, [[height-1],[width-1]] ))
fixed_image[-1,-1] = med
return hot_pixels,fixed_image
hot_pixels,fixed_image = find_outlier_pixels(Z)
for y,x in zip(hot_pixels[0],hot_pixels[1]):
ax1.plot(x,y,'ro',mfc='none',mec='r',ms=10)
ax1.set_xlim(0,200)
ax1.set_ylim(0,200)
ax2.set_title('Image with hot pixels removed')
ax2.imshow(fixed_image,interpolation='nearest',origin='lower',clim=(0,255))
plt.show()
输出:
关于python - 自动从 python 中的图像中删除热/死像素,我们在Stack Overflow上找到一个类似的问题: https://stackoverflow.com/questions/18951500/